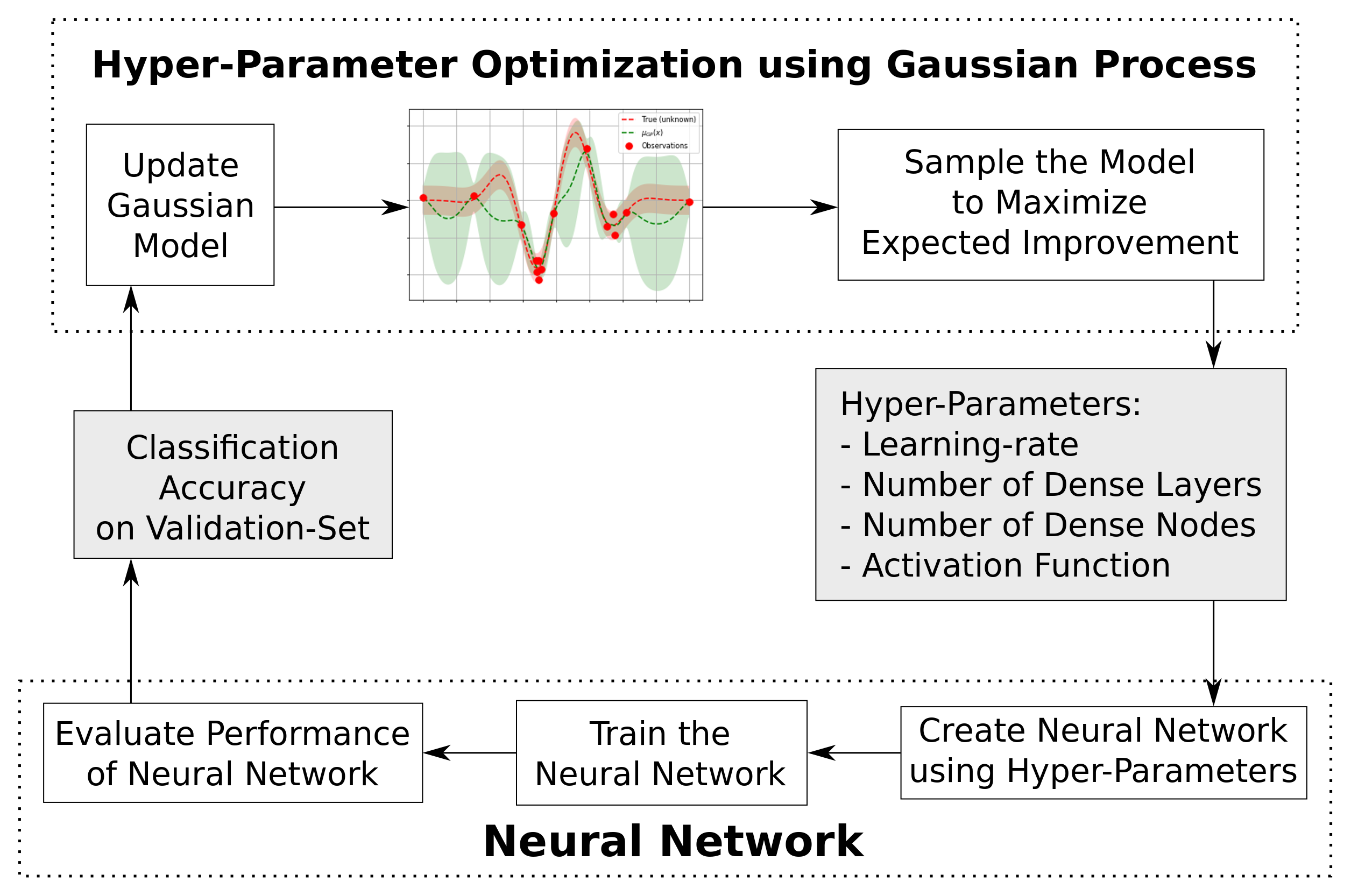
Hyper Parameter Tuning
Hyper Parameter Tuning
- One way of searching for good hyper-parameters is by hand-tuning
- Another way of searching for good hyper-parameters is to divide each parameter’s valid range into evenly spaced values, and then simply have the computer try all combinations of parameter-values. This is called Grid Search.
- another way of searching for good hyper-parameters is by random search.
- This tutorial uses a clever method for finding good hyper-parameters known as Bayesian Optimization
Basic library from scikit-optimize
import skopt
from skopt import gp_minimize, forest_minimize
from skopt.space import Real, Categorical, Integer
from skopt.plots import plot_convergence
from skopt.plots import plot_objective, plot_evaluations
# from skopt.plots import plot_histogram, plot_objective_2D
from skopt.utils import use_named_args
Hyper parameter
dim_learning_rate = Real(low=1e-6, high=1e-2, prior='log-uniform',
name='learning_rate')
dim_num_dense_layers = Integer(low=1, high=5, name='num_dense_layers')
dim_num_dense_nodes = Integer(low=5, high=512, name='num_dense_nodes')
dim_activation = Categorical(categories=['relu', 'sigmoid'],
name='activation')
dimensions = [dim_learning_rate,
dim_num_dense_layers,
dim_num_dense_nodes,
dim_activation]
Fitness Function
This is the function that creates and trains a neural network with the given hyper-parameters, and then evaluates its performance on the validation-set. The function then returns the so-called fitness value (aka. objective value), which is the negative classification accuracy on the validation-set. It is negative because skopt performs minimization instead of maximization.
Note the function decorator @use_named_args which wraps the fitness function so that it can be called with all the parameters as a single list, for example: fitness(x=[1e-4, 3, 256, 'relu']). This is the calling-style skopt uses internally.
@use_named_args(dimensions=dimensions)
def fitness(learning_rate, num_dense_layers,
num_dense_nodes, activation):
"""
Hyper-parameters:
learning_rate: Learning-rate for the optimizer.
num_dense_layers: Number of dense layers.
num_dense_nodes: Number of nodes in each dense layer.
activation: Activation function for all layers.
"""
# Print the hyper-parameters.
print('learning rate: {0:.1e}'.format(learning_rate))
print('num_dense_layers:', num_dense_layers)
print('num_dense_nodes:', num_dense_nodes)
print('activation:', activation)
print()
# Create the neural network with these hyper-parameters.
model = create_model(learning_rate=learning_rate,
num_dense_layers=num_dense_layers,
num_dense_nodes=num_dense_nodes,
activation=activation)
# Dir-name for the TensorBoard log-files.
log_dir = log_dir_name(learning_rate, num_dense_layers,
num_dense_nodes, activation)
# Create a callback-function for Keras which will be
# run after each epoch has ended during training.
# This saves the log-files for TensorBoard.
# Note that there are complications when histogram_freq=1.
# It might give strange errors and it also does not properly
# support Keras data-generators for the validation-set.
callback_log = TensorBoard(
log_dir=log_dir,
histogram_freq=0,
batch_size=32,
write_graph=True,
write_grads=False,
write_images=False)
# Use Keras to train the model.
history = model.fit(x=data.train.images,
y=data.train.labels,
epochs=3,
batch_size=128,
validation_data=validation_data,
callbacks=[callback_log])
# Get the classification accuracy on the validation-set
# after the last training-epoch.
accuracy = history.history['val_acc'][-1]
# Print the classification accuracy.
print()
print("Accuracy: {0:.2%}".format(accuracy))
print()
# Save the model if it improves on the best-found performance.
# We use the global keyword so we update the variable outside
# of this function.
global best_accuracy
# If the classification accuracy of the saved model is improved ...
if accuracy > best_accuracy:
# Save the new model to harddisk.
model.save(path_best_model)
# Update the classification accuracy.
best_accuracy = accuracy
# Delete the Keras model with these hyper-parameters from memory.
del model
# Clear the Keras session, otherwise it will keep adding new
# models to the same TensorFlow graph each time we create
# a model with a different set of hyper-parameters.
K.clear_session()
# NOTE: Scikit-optimize does minimization so it tries to
# find a set of hyper-parameters with the LOWEST fitness-value.
# Because we are interested in the HIGHEST classification
# accuracy, we need to negate this number so it can be minimized.
return -accuracy
Run the Hyper-Parameter Optimization
search_result = gp_minimize(func=fitness,
dimensions=dimensions,
acq_func='EI', # Expected Improvement.
n_calls=40,
x0=default_parameters)
Result
plot_convergence(search_result)
search_result.x
space.point_to_dict(search_result.x)
search_result.fun
sorted(zip(search_result.func_vals, search_result.x_iters))
fig, ax = plot_histogram(result=search_result,
dimension_name='activation')
Scikit-optimize
from sklearn.base import clone
from skopt import gp_minimize
from skopt.learning import GaussianProcessRegressor
from skopt.learning.gaussian_process.kernels import ConstantKernel, Matern
# Use custom kernel and estimator to match previous example
m52 = ConstantKernel(1.0) * Matern(length_scale=1.0, nu=2.5)
gpr = GaussianProcessRegressor(kernel=m52, alpha=noise**2)
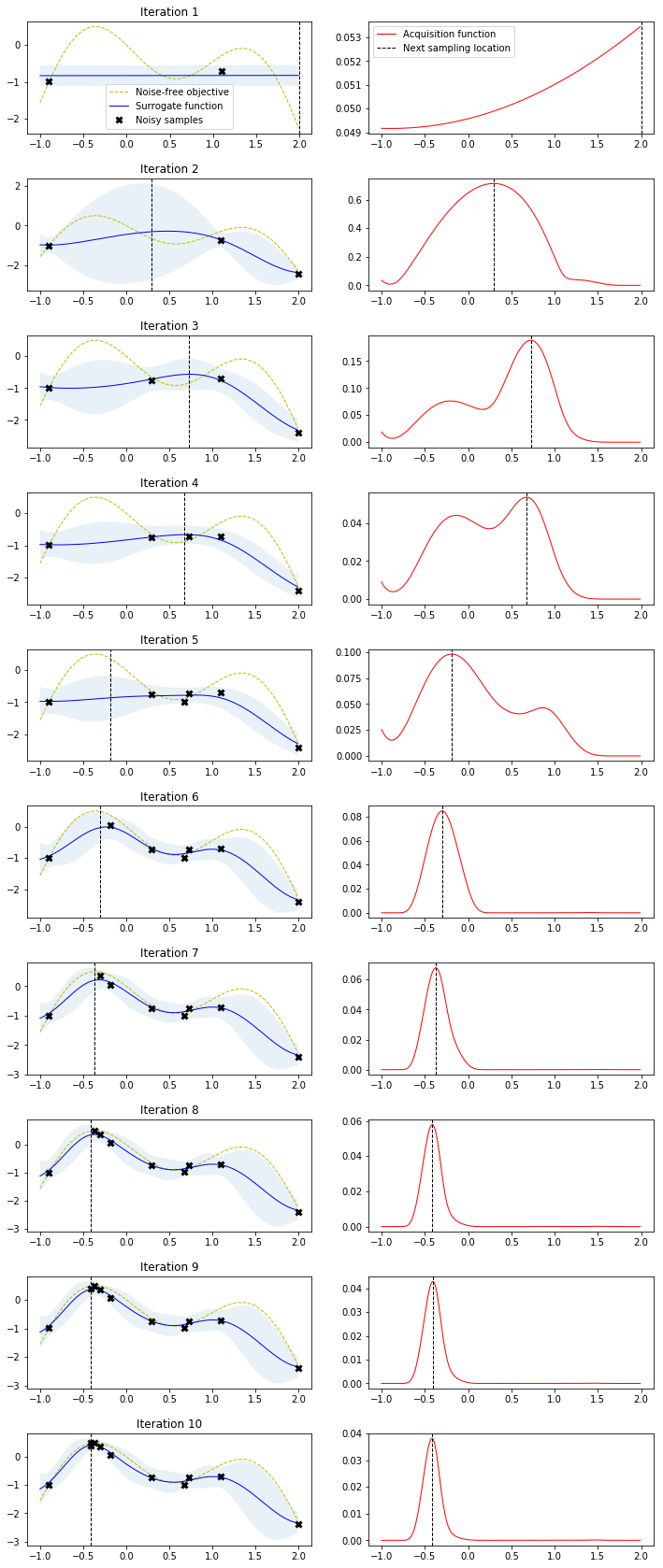
r = gp_minimize(lambda x: -f(np.array(x))[0],
bounds.tolist(),
base_estimator=gpr,
acq_func='EI', # expected improvement
xi=0.01, # exploitation-exploration trade-off
n_calls=10, # number of iterations
n_random_starts=0, # initial samples are provided
x0=X_init.tolist(), # initial samples
y0=-Y_init.ravel())
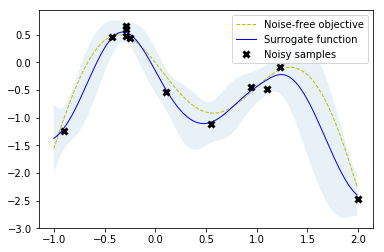
# Fit GP model to samples for plotting results
gpr.fit(r.x_iters, -r.func_vals)
# Plot the fitted model and the noisy samples
plot_approximation(gpr, X, Y, r.x_iters, -r.func_vals, show_legend=True)
GPyOpt
import GPy
import GPyOpt
from GPyOpt.methods import BayesianOptimization
kernel = GPy.kern.Matern52(input_dim=1, variance=1.0, lengthscale=1.0)
bds = [{'name': 'X', 'type': 'continuous', 'domain': bounds.ravel()}]
optimizer = BayesianOptimization(f=f,
domain=bds,
model_type='GP',
kernel=kernel,
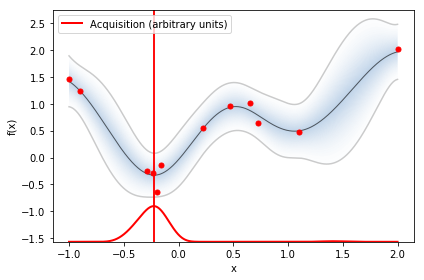
acquisition_type ='EI',
acquisition_jitter = 0.01,
X=X_init,
Y=-Y_init,
noise_var = noise**2,
exact_feval=False,
normalize_Y=False,
maximize=True)
optimizer.run_optimization(max_iter=10)
optimizer.plot_acquisition()