XGBoost
Example XGboost Grid Search in Python
- XGBoost (Extreme Gradient Boosting) belongs to a family of boosting algorithms and uses the gradient boosting (GBM) framework at its core. It is an optimized distributed gradient boosting library.
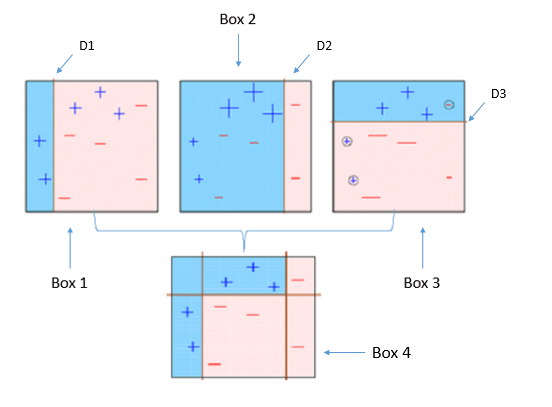
- Box 4: This is a weighted combination of the weak classifiers (Box 1,2 and 3). As you can see, it does a good job at classifying all the points correctly.
XGBoost’s hyperparameters
- learning_rate: step size shrinkage used to prevent overfitting. Range is [0,1]
- max_depth: determines how deeply each tree is allowed to grow during any boosting round.
- subsample: percentage of samples used per tree. Low value can lead to underfitting.
- colsample_bytree: percentage of features used per tree. High value can lead to overfitting.
- n_estimators: number of trees you want to build.
- objective: determines the loss function to be used like reg:linear for regression problems, reg:logistic for classification problems with only decision,
-
binary:logistic for classification problems with probability.
- gamma: controls whether a given node will split based on the expected reduction in loss after the split. A higher value leads to fewer splits. Supported only for tree-based learners.
- alpha: L1 regularization on leaf weights. A large value leads to more regularization.
- lambda: L2 regularization on leaf weights and is smoother than L1 regularization.
def train(self):
params = {}
params['objective'] = 'multi:softprob'
params['eta'] = 0.01
params['num_class'] = 2
params['max_depth'] = 20
params['subsample'] = 0.05
params['colsample_bytree'] = 0.05
params['eval_metric'] = 'mlogloss'
#params['scale_pos_weight'] = 10
#params['silent'] = True
#params['gpu_id'] = 0
#params['max_bin'] = 16
#params['tree_method'] = 'gpu_hist'
train = xgb.DMatrix(self.data, self.labels)
test = xgb.DMatrix(self.test_data, self.test_labels)
watchlist = [(train, 'train'), (test, 'test')]
clf = xgb.train(params, train, 1000, evals=watchlist, early_stopping_rounds=100)
joblib.dump(clf, 'models/clf.pkl')
cm = confusion_matrix(self.test_labels, list(map(lambda x: int(x[1] > .5), clf.predict(test))))
# # print("self.test_labels {}".format(self.test_labels))
# global predict_result
# predict_result = clf.predict(test)
# global test_labels
# test_labels = self.test_labels
print(cm)
plot_confusion_matrix(cm, ['Down', 'Up'], normalize=True, title="Confusion Matrix")